Ammar Ammar in my group just published the second half of his cheminformatics study into what happens with binding affinities when the proteins show amino acid changes, selected based on world-wide population statistics. His idea what that drugs should be designed to not be selective for a particular genotype. The first paper (see this post) tells the story about how to automate running thousands of docking experiments and explains how to put this knowledge base online, while the paper this month explains how machine learning can learn the patterns found in those docking experiments:
- 2022: PSnpBind: a database of mutated binding site protein–ligand complexes constructed using a multithreaded virtual screening workflow
- 2023: PSnpBind-ML: predicting the effect of binding site mutations on protein-ligand binding affinity
The idea in the PSnpBind-ML paper is simple. We can calculate binding affinities for many ligand-protein complexes. If we calculate enough, one can create a QSAR model that includes ligand and protein information (to capture the SNP uniqueness) to predict that affinity with the QSAR model. That will be a lot more scalable. The 2023 article has the full details, and everything is open.
One thing that fascinated me when Ammar proposed this study is the notion that in this way, for each ligand, we can see how stable the binding affinity is over the various protein variants. Are there proteins which are harder to target because of the variants? Do certain classes of chemical structures show a lot of binding differences over the world-wide protein diversity?
Also, I was interested in if it would work in the first place. I remember from my own PhD days (some 20 years ago now) that docking experiments had a fairly high prediction error. So, when I see this plot on the independent test set, I am intrigued:
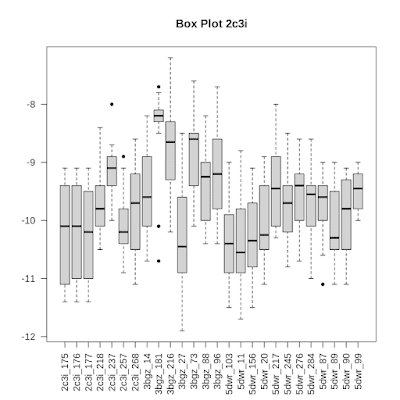
So, what about the binding affinity variation? Ammar did not put this figure in the paper, but sent me a copy to put online here.
We here see a boxplot (yeah, there are better alternatives, I know...) showing quite a bit of variation in the binding affinities for the various variants of human Pim-1 kinase (with crystal structures 2C3I, 3BGZ, etc). These plots show that the variation is mostly high, but sometimes quite small indeed. I don't really see a pattern here.
And totally in line with open science, each combination of docked ligand and mutated protein can be looked at online with Jmol, e.g. this one:
In this case, the amino acid change is right next to the ligand. Ammar selected them as such. Of course, biology in reality is much more complex. And maybe the differences we see here are not even significant compared to other effects.
But one thing keeps wondering, and I hope someone can explain this to me, in the past I would see experimental data on ligand-protein binding referring to the protein, but not so much the protein variant. We would need a lot of experimental measurements of ligands binding to protein variants to validate this.
But all this uncertainty of the biological and drug discovery implications, there is another reason why I am really happy about this story. First, the openness and the ability to share it FAIR-ly online (check his use of w3id, e.g. https://w3id.org/psnpbind/protein/2c3i), and, second, the notion that we can do things now at this scale. With all the deep learning discussions ongoing, the ability to inspect in detail what these models do, how they behave, the "explainable AI" if you like, is essential and Ammar showed here how to do that.
Thinking back about the study about pKa's of warfarin tautomers, being all over the place from very basic to very acidic, it is nice to see some data on the effect of the SNPs on the binding affinities.
I am sure you have some thoughts on this work. We did ask someone about the idea before we started, and we were told it had limited use. Use the comment section, or better even, write a reply blog post on your own platform, or send us an email. Looking forward to hearing from you.


No comments:
Post a Comment